 MemMoRF database of membrane
associated disordered protein
regions
MemMoRF database of membrane
associated disordered protein
regions
Membrane-binding Molecular Recognition Features
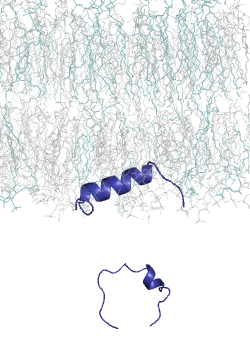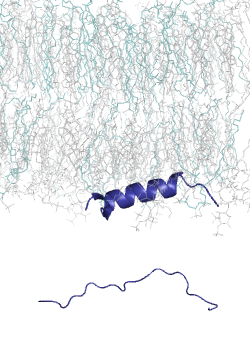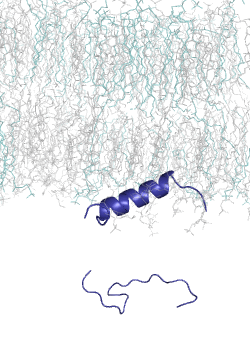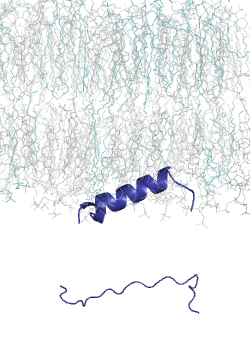
MemMoRF is a comprehensive database of transmembrane and membrane associated proteins with MemMoRFs: regions that undergo disorder-to-order transition or large conformational changes upon membrane association. These proteins play essential roles in various biological processes and participate in signaling, regulation of membrane transport, and viral invasion. The MemMoRF regions correspond to important molecular switches which form highly regulated, dynamic interactions with specific membranes and other proteins. Importantly, a large fraction of these proteins are associated with disease causing mutations (e.g. prolactin receptor, EGFR, Notch ligand DLL4, and sodium/potassium ATPase subunit γ) and some of the mutations are specifically localized inside memMoRF segments (e.g. in the case of integrins, phospholamban, the voltage-gated potassium channel KCNE1, and proteins associated with neurodegenerative diseases). MemMoRFs represent desirable therapeutic targets, however, their modulation is currently challenging due to their membrane attachment and flexible nature.
MemMoRFs were collected from the literature through manual curation. As data on these membrane interacting regions are scattered in the literature, efforts were directed towards transmembrane and peripheral membrane proteins with NMR structures, which were collected from the UniProt and PDB databases, respectively. We also explored static structures and investigated invisible regions of X-ray and cryo-EM structures, which belong to transmembrane or membrane associated proteins possessing annotated disordered regions. Papers accompanying the collected structures were manually searched for disordered regions with mentions of membrane interactions. The disorder property of the structures was also characterized by calculating secondary structure populations and residue flexibility based on chemical shifts from BMRB, using δ2D and RCI, respectively.
Entries in the MemMoRF database can be categorized based on the type of the lipid binding induced conformational change, the availability of NMR structures, the location relative to the membrane and the presence of disease mutations. The database currently contains 107 proteins and 149 membrane interacting regions.