Protein-membrane interactions play fundamental roles in virtually every aspect of cellular function. It is increasingly recognized that such interactions often rely on intrinsically disordered regions (IDRs) that can form highly specific reversible interactions not only with other proteins but also with biological membranes. In contrast to IDRs that exhibit disorder-order transition upon interaction with protein partners (called Molecular Recognition Features or MoRFs, PMID: 14749181, PMID: 16935303), segments involved in lipid induced alteration of order/disorder state (termed MemMoRFs) represent a distinct category of context-dependent behavior.
The database stores information on close to one hundred MemMoRF segments and on associated proteins. The entries were manually collected and curated.
Our web application provides:
- cross-reference to UniProt entries
- sequence boundaries of MemMoRF regions
- type of transition upon membrane bilayer binding
- structural data (PDB, BMRB)
- hydrophobicity plot, IUPred short predictions
- membrane mimetic environment of NMR experiments if available
- literature references from PubMed
- disease associated data and cross-references to MIM and dbSNP
- protein-protein interaction network
We screened NMR structures of transmembrane proteins (TMPs) and membrane-associated proteins (MAPs) for MemMoRFs. We put emphasis to exploit experimental information, which was derived from the literature and structures determined by NMR, for MemMoRF identification. To characterize the transient secondary structures of IDRs, we did not rely solely on the deposited NMR structures, but also incorporated secondary structure propensity (δ2D, PMID: 22360139) and random coil index (RCI, PMID: 16248604) calculated directly from chemical shifts. We also investigated invisible regions of X-ray and cryo-EM structures, which belong to transmembrane or membrane associated proteins annotated in DisProt.
For being a MemMoRF, two requirements need to be satisfied:
- The region has to be located in an IDR; the associated information may originate from
- papers: the region was mentioned as an IDR in papers
- calculations: (a) “coil” secondary structure population and high flexibility from NMR experiments in the absence of a membrane mimetic; (b) flexible non-coiled region within a highly flexible coil region in the presence of a membrane mimetics
- databases: the region is presented as an IDR in DisProt, DIBS or PFAM
- The region has to be mentioned to interact with membranes in the literature (annotated as “papers” proof). Complementary information may originate from
- calculations: an NMR experiment shows non-coiled secondary structure population for the region of interest, in the presence of membrane mimetics
- structures: membrane association is supported by structural information (e.g. the juxtamembrane region is perpendicular to the membrane plane as in the case of Phospholemman)
Based on proofs from the above mentioned data sources, a membrane-interacting non-transmembrane region can be categorized as ordered (order-to-order transition upon bilayer binding) or MemMoRFs. We observed all three possible combinations for transitions associated to lipid binding, such as disorder-to-order, disorder-to-disorder, and order-to-disorder transitions.
Proteins containing MemMoRFs or ordered membrane-interacting regions are presented in a searchable and sortable table in the Browse page. The table shows basic information from UniProt about each protein (name, gene name, Organism, UniProt Accession, and keywords), as well as the following columns:
- Type of hits in the protein (disorder to order, disorder to disorder, order to disorder, bistable secondary structure)
- Whether there are relevant structures associated with the entry page
- Whether the protein is annotated as transmembrane by UniProt or not
- Whether δ2D and RCI calculations are available for the protein
- Whether there are disease phenotypes from MIM available
The Entry page repeats basic information about the protein.
The next table summarizes collected MemMoRF data
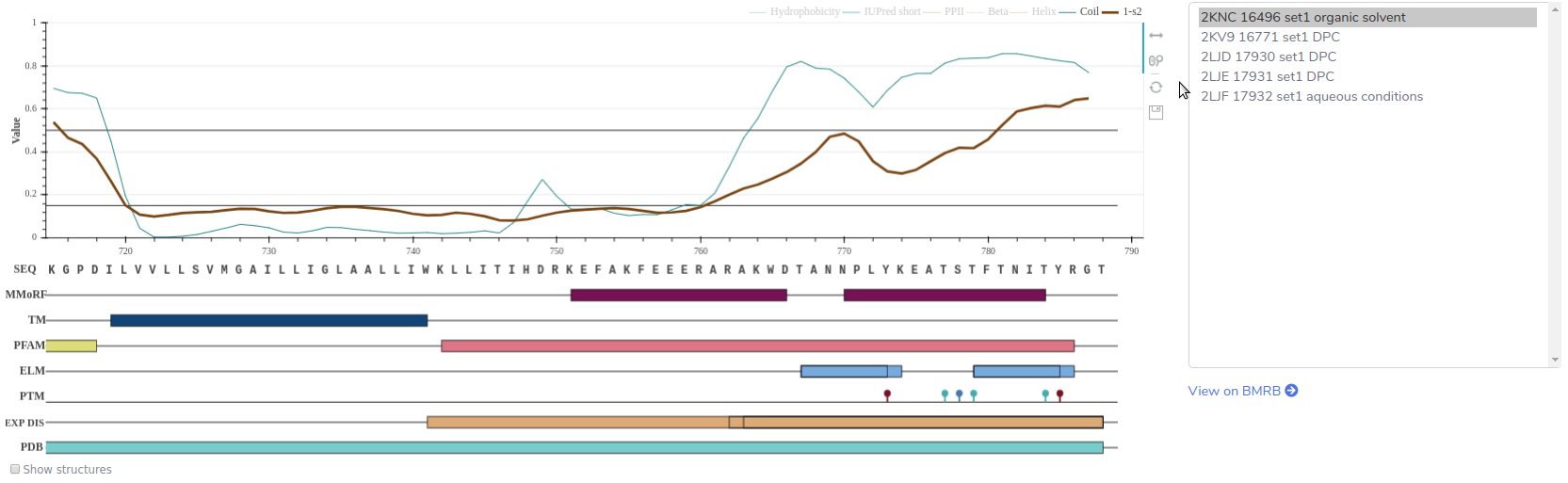
The main graph, which can be saved using the controls next to it, includes the δ2D, RCI, hydrophobicity, and IUPred short calculations, as well as sequence specific data, such as location of TM helices, PFAM domains, ELM, PTM, disordered regions and segments covered in the Protein Data Bank. Thresholds of δ2D and RCI calculations are also shown at 0.5 and 0.15 values, respectively. If several NMR experiments are associated with the entry, the user can select one from a box, right from the graph. This box lists the membrane mimetics, if used in the NMR experiment, the PDB ID of the calculated NMR ensemble, the BMRB ID, and the “_Assigned_chem_shift_list.ID” of BMRB used for the calculation.
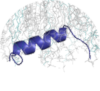
Structures strongly associated with the MemMoRF can be selected for display and manipulation (e.g. coloring and rotation) using LiteMol.
In order to assess the role of the entry (MemMoRF and protein) in pathophysiological states and in the context of their protein interacting partners, the following information are listed and linked to the source in the bottom region of the entry page:
- Disease phenotypes from MIM if available
- Disease causing and polymorphic variations from dbSNP
- Drug Bank record of the protein
- Protein-protein interactions with link to the IntAct entry of the protein and both link and an interactive, embedded network graph from STRING
Various statistics are available, such as on MemMoRF types and organisms at the Statistics page.
Data can be downloaded in JSON, TSV, and XML formats.
Individual entries can be accessed through our REST API, such as http://memmorf.hegelab.org/UNIPROT_ACC.json. For details and an example script please see the Download page.