Integrin beta-3 (P05106)
Basic protein information
UniProt Gene
ITGB3
Organism
Homo sapiens
Keywords
3D-structure, Alternative splicing, Cell adhesion, Cell junction, Cell membrane, Cell projection, Direct protein sequencing, Disease mutation, Disulfide bond, Glycoprotein, Host cell receptor for virus entry, Host-virus interaction, Integrin, Membrane, Phosphoprotein, Polymorphism, Postsynaptic cell membrane, Receptor, Reference proteome, Repeat, Signal, Synapse, Transmembrane, Transmembrane helix
MemMoRF data
Warning: residue numbering from UniProt, PDB, and papers may differ.
| MemMoRF ID | Region | Supporting statements | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 9228 | 770-784 |
|
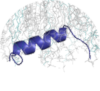
"Immediately next to the loop is the N744PLY747 motif that forms an inverse turn, which is the start point for the second helix" ... "The formation of the C-terminal helix is surprising, because it was absent in the structure of the αIIb/β3 tail complex in aqueous solution. Because no long-range NOEs were observed between the N- and C-terminal helices, the formation of the C-terminal helix is likely induced and stabilized by the docking of the NPLY turn onto DPC." (PMID:15024114) "As expected, TMproxH was predicted to partition into the bilayer perpendicular to the membrane as an extension of the TM helix, whereas CytoH1 and CytoH2 were predicted to lie along the membrane surface, stabilized by hydrophobic residues Phe727 and Phe730 in CytoH1 and Leu746, Tyr747, Ala750, Phe754, and Ile757 in CytoH2" ... "HDX of the β3 cytoplasmic domain conjugated to lipid." ... "Asn743-Thr762 was not observed in the absence the bilayers [fig4]" ... Results of HDX experiments (Admin): "the intermediate fragment spanning a portion of the loop and all of CytoH2 had four protected amides (Asn743-Thr762); and the most C-terminal fragment (Asn756-Thr762) had only one protected amide. These results confirmed the location of the helices identified by NMR, as well as the hierarchy of increasing disorder that progresses from the N terminus to the C terminus of the β3 cytoplasmic domain." (PMID:21156831) "Moreover, we have demonstrated that several residues of β3NP (Trp739, Thr741, Ala742, Pro745, and Tyr747) could interact with DPC micelles and these interactions initiate the formation of a second short α-helical region (Leu746-Asn756), which is not generally observed in either aqueous β3 or αIIbβ3 heterodimer" ... ". In this study, we also have accumulated the first direct evidence that tyrosine phosphorylation affects the structure and the association of 3CT with membrane." (PMID:21956114) "At the TM-CT border, the spatial positions of the positively charged groups of αIIb K989/β3 K716 (Fig. 1D) suggest that they begin the cytoplasmic regions. However, it is possible that the side chains following membrane-proximal residues αIIb V990-F993, β3 L717-I721 are inserted into or anchored onto the membrane (Fig. 1D), which may stabilize the relative orientations of the TM helices." (PMID:19805198) |
||||||||||
| 9227 | 751-766 |
|
"As expected, TMproxH was predicted to partition into the bilayer perpendicular to the membrane as an extension of the TM helix, whereas CytoH1 and CytoH2 were predicted to lie along the membrane surface, stabilized by hydrophobic residues Phe727 and Phe730 in CytoH1 and Leu746, Tyr747, Ala750, Phe754, and Ile757 in CytoH2" ... "HDX of the β3 cytoplasmic domain conjugated to lipid." ... "Asn743-Thr762 was not observed in the absence the bilayers [fig4]" (PMID:21156831) "Moreover, we have demonstrated that several residues of β3NP (Trp739, Thr741, Ala742, Pro745, and Tyr747) could interact with DPC micelles and these interactions initiate the formation of a second short α-helical region (Leu746-Asn756), which is not generally observed in either aqueous β3 or αIIbβ3 heterodimer" ... ". In this study, we also have accumulated the first direct evidence that tyrosine phosphorylation affects the structure and the association of 3CT with membrane." (PMID:21956114) |
Strutures
Genetic
variations/dbSNP
| a.a. change | Type | dbSNP ID | Disesase name | |
|---|---|---|---|---|
| p.Ser778Pro | Disease | within MemMoRF region | 121918447 | Glanzmann |
| p.Leu59Pro | Polymorphism | 5918 | - | |
| p.Cys64Tyr | Disease | 74554539 | Glanzmann | |
| p.Leu66Arg | Polymorphism | 36080296 | - | |
| p.Arg119Trp | Disease | 781062792 | Glanzmann | |
| p.Tyr141Cys | Disease | - | Glanzmann | |
| p.Leu143Trp | Disease | 121918452 | Glanzmann | |
| p.Met144Arg | Disease | 77963874 | Glanzmann | |
| p.Asp145Tyr | Disease | 121918445 | Glanzmann | |
| p.Asp145Asn | Disease | - | Glanzmann | |
| p.Met150Val | Disease | 767548512 | Glanzmann | |
| p.Thr166Ile | Polymorphism | 74708909 | - | |
| p.Arg169Gln | Polymorphism | 5917 | - | |
| p.Ser188Leu | Disease | 143146734 | Glanzmann | |
| p.Leu222Pro | Disease | 79208797 | Glanzmann | |
| p.Arg240Gln | Disease | 121918444 | Glanzmann | |
| p.Arg240Trp | Disease | 121918446 | Glanzmann | |
| p.Arg242Gln | Disease | 377162158 | Glanzmann | |
| p.Asp243Val | Disease | - | Glanzmann | |
| p.Gly247Asp | Disease | 79560904 | Glanzmann | |
| p.Lys279Met | Disease | 79775494 | Glanzmann | |
| p.Leu288Pro | Disease | - | Glanzmann | |
| p.His306Pro | Disease | 13306476 | Glanzmann | |
| p.Met321Leu | Disease | - | Glanzmann | |
| p.Ile330Asn | Disease | - | Glanzmann | |
| p.Cys400Tyr | Disease | 121918449 | Glanzmann | |
| p.Pro433Ala | Polymorphism | 121918448 | - | |
| p.Val453Ile | Polymorphism | 5921 | - | |
| p.Arg515Gln | Polymorphism | 13306487 | - | |
| p.Cys532Tyr | Disease | - | Glanzmann | |
| p.Cys568Arg | Disease | - | Glanzmann | |
| p.Cys586Phe | Disease | - | Glanzmann | |
| p.Cys586Arg | Disease | - | Glanzmann | |
| p.Gly598Ser | Disease | - | Glanzmann | |
| p.Cys601Arg | Disease | 747534508 | Glanzmann | |
| p.Gly605Ser | Disease | 144884023 | Glanzmann | |
| p.Arg662Cys | Polymorphism | 151219882 | - | |
| p.Asp749His | Disease | 398122372 | Bleeding |
Protein-protein interactions
Click on a node for details.
v. 1.0

 http://www.ebi.ac.uk/intact/query/P05106
http://www.ebi.ac.uk/intact/query/P05106
